FASTA
| Developer(s) | |
|---|---|
| Stable release | 36
|
| Repository | |
| Operating system | |
| Type | Bioinformatics |
| License | apache2.0 |
| Website |
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985.[1] Its legacy is the FASTA format which is now ubiquitous in bioinformatics.
History[edit]
The original FASTA program was designed for protein sequence similarity searching. Because of the exponentially expanding genetic information and the limited speed and memory of computers in the 1980s heuristic methods were introduced aligning a query sequence to entire data-bases. FASTA, published in 1987, added the ability to do DNA:DNA searches, translated protein:DNA searches, and also provided a more sophisticated shuffling program for evaluating statistical significance.[2] There are several programs in this package that allow the alignment of protein sequences and DNA sequences. Nowadays, increased computer performance makes it possible to perform searches for local alignment detection in a database using the Smith–Waterman algorithm.
FASTA is pronounced "fast A", and stands for "FAST-All", because it works with any alphabet, an extension of the original "FAST-P" (protein) and "FAST-N" (nucleotide) alignment tools.

Uses[edit]
The current FASTA package contains programs for protein:protein, DNA:DNA, protein:translated DNA (with frameshifts), and ordered or unordered peptide searches. Recent versions of the FASTA package include special translated search algorithms that correctly handle frameshift errors (which six-frame-translated searches do not handle very well) when comparing nucleotide to protein sequence data.
In addition to rapid heuristic search methods, the FASTA package provides SSEARCH, an implementation of the optimal Smith–Waterman algorithm.
A major focus of the package is the calculation of accurate similarity statistics, so that biologists can judge whether an alignment is likely to have occurred by chance, or whether it can be used to infer homology. The FASTA package is available from the University of Virginia[3] and the European Bioinformatics Institute.[4]
The FASTA file format used as input for this software is now largely used by other sequence database search tools (such as BLAST) and sequence alignment programs (Clustal, T-Coffee, etc.).
Search method[edit]
FASTA takes a given nucleotide or amino acid sequence and searches a corresponding sequence database by using local sequence alignment to find matches of similar database sequences.
The FASTA program follows a largely heuristic method which contributes to the high speed of its execution. It initially observes the pattern of word hits, word-to-word matches of a given length, and marks potential matches before performing a more time-consuming optimized search using a Smith–Waterman type of algorithm.
The size taken for a word, given by the parameter kmer, controls the sensitivity and speed of the program. Increasing the k-mer value decreases number of background hits that are found. From the word hits that are returned the program looks for segments that contain a cluster of nearby hits. It then investigates these segments for a possible match.
There are some differences between fastn and fastp relating to the type of sequences used but both use four steps and calculate three scores to describe and format the sequence similarity results. These are:
- Identify regions of highest density in each sequence comparison. Taking a k-mer to equal 1 or 2.
- In this step all or a group of the identities between two sequences are found using a look up table. The k-mer value determines how many consecutive identities are required for a match to be declared. Thus the lesser the k-mer value: the more sensitive the search. k-mer=2 is frequently taken by users for protein sequences and kmer=4 or 6 for nucleotide sequences. Short oligonucleotides are usually run with k-mer= 1. The program then finds all similar local regions, represented as diagonals of a certain length in a dot plot, between the two sequences by counting k-mer matches and penalizing for intervening mismatches. This way, local regions of highest density matches in a diagonal are isolated from background hits. For protein sequences BLOSUM50 values are used for scoring k-mer matches. This ensures that groups of identities with high similarity scores contribute more to the local diagonal score than to identities with low similarity scores. Nucleotide sequences use the identity matrix for the same purpose. The best 10 local regions selected from all the diagonals put together are then saved.
- Rescan the regions taken using the scoring matrices. trimming the ends of the region to include only those contributing to the highest score.
- Rescan the 10 regions taken. This time use the relevant scoring matrix while rescoring to allow runs of identities shorter than the k-mer value. Also while rescoring conservative replacements that contribute to the similarity score are taken. Though protein sequences use the BLOSUM50 matrix, scoring matrices based on the minimum number of base changes required for a specific replacement, on identities alone, or on an alternative measure of similarity such as PAM, can also be used with the program. For each of the diagonal regions rescanned this way, a subregion with the maximum score is identified. The initial scores found in step1 are used to rank the library sequences. The highest score is referred to as init1 score.
- In an alignment if several initial regions with scores greater than a CUTOFF value are found, check whether the trimmed initial regions can be joined to form an approximate alignment with gaps. Calculate a similarity score that is the sum of the joined regions penalising for each gap 20 points. This initial similarity score (initn) is used to rank the library sequences. The score of the single best initial region found in step 2 is reported (init1).
- Here the program calculates an optimal alignment of initial regions as a combination of compatible regions with maximal score. This optimal alignment of initial regions can be rapidly calculated using a dynamic programming algorithm. The resulting score initn is used to rank the library sequences. This joining process increases sensitivity but decreases selectivity. A carefully calculated cut-off value is thus used to control where this step is implemented, a value that is approximately one standard deviation above the average score expected from unrelated sequences in the library. A 200-residue query sequence with k-mer 2 uses a value 28.
- Use a banded Smith–Waterman algorithm to calculate an optimal score for alignment.
- This step uses a banded Smith–Waterman algorithm to create an optimised score (opt) for each alignment of query sequence to a database(library) sequence. It takes a band of 32 residues centered on the init1 region of step2 for calculating the optimal alignment. After all sequences are searched the program plots the initial scores of each database sequence in a histogram, and calculates the statistical significance of the "opt" score. For protein sequences, the final alignment is produced using a full Smith–Waterman alignment. For DNA sequences, a banded alignment is provided.
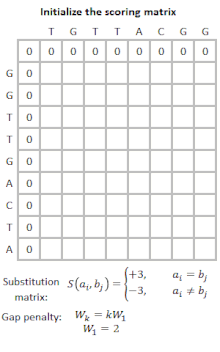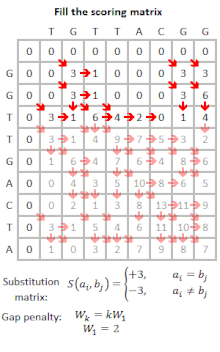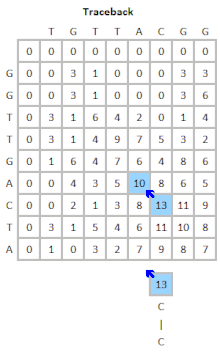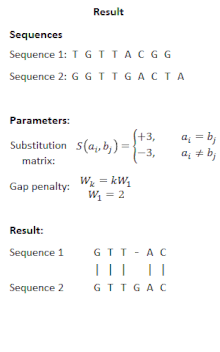
FASTA can remove complexity regions before aligning the sequences by encoding low complexity regions in lower case and using the -S option. However, the BLAST program offers more options for correcting for biased composition statistics. Therefore, the program PRSS is added in the FASTA distribution package. PRSS shuffles the matching sequences in the database either on the one-letter level or it shuffles short segments which length the user can determine. The shuffled sequences are now aligned again and if the score is still higher than expected this is caused by the low complexity regions being mixed up still mapping to the query. By the amount of the score the shuffled sequences still attain PRSS now can predict the significance of the score of the original sequences. The higher the score of the shuffled sequences the less significant the matches found between original database and query sequence.[5]
The FASTA programs find regions of local or global similarity between Protein or DNA sequences, either by searching Protein or DNA databases, or by identifying local duplications within a sequence. Other programs provide information on the statistical significance of an alignment. Like BLAST, FASTA can be used to infer functional and evolutionary relationships between sequences as well as help identify members of gene families.
See also[edit]
References[edit]
- ^ Lipman, DJ; Pearson, WR (1985). "Rapid and sensitive protein similarity searches". Science. 227 (4693): 1435–41. Bibcode:1985Sci...227.1435L. doi:10.1126/science.2983426. PMID 2983426.

- ^ Pearson, WR; Lipman, DJ (1988). "Improved tools for biological sequence comparison". Proceedings of the National Academy of Sciences of the United States of America. 85 (8): 2444–8. Bibcode:1988PNAS...85.2444P. doi:10.1073/pnas.85.8.2444. PMC 280013. PMID 3162770.
- ^ "FASTA Programs". Archived from the original on 2000-03-04.
- ^ "FASTA/SSEARCH/GGSEARCH/GLSEARCH < Sequence Similarity Searching < EMBL-EBI".
- ^ David W. Mount: Bioinformatics Sequence and Genome Analysis, Edition 1, Cold Spring Harbor Laboratory Press, 2001, pp. 295–297.

